Homework 2
1.
|
|
Gly |
Ala |
Val |
Leu |
Ile |
Met |
Cys |
Ser |
Thr |
Asn |
Gln |
Asp |
Glu |
Lys |
Arg |
His |
Phe |
Tyr |
Trp |
Pro |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Gly |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Ala |
58 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Val |
10 |
37 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Leu |
2 |
10 |
30 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Ile |
|
7 |
66 |
25 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Met |
1 |
3 |
8 |
21 |
6 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Cys |
1 |
3 |
3 |
|
2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Ser |
45 |
77 |
4 |
3 |
2 |
2 |
12 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Thr |
5 |
59 |
19 |
5 |
13 |
3 |
1 |
70 |
|
|
|
|
|
|
|
|
|
|
|
|
|
Asn |
16 |
11 |
1 |
4 |
4 |
|
|
43 |
17 |
|
|
|
|
|
|
|
|
|
|
|
|
Gln |
3 |
9 |
3 |
8 |
1 |
2 |
|
5 |
4 |
5 |
|
|
|
|
|
|
|
|
|
|
|
Asp |
16 |
15 |
2 |
|
1 |
|
|
10 |
6 |
53 |
8 |
|
|
|
|
|
|
|
|
|
|
Glu |
11 |
27 |
4 |
2 |
4 |
1 |
|
9 |
3 |
9 |
42 |
83 |
|
|
|
|
|
|
|
|
|
Lys |
6 |
6 |
2 |
4 |
4 |
9 |
|
17 |
20 |
32 |
15 |
|
10 |
|
|
|
|
|
|
|
|
Arg |
1 |
3 |
2 |
2 |
3 |
2 |
1 |
14 |
2 |
2 |
12 |
9 |
|
48 |
|
|
|
|
|
|
|
His |
1 |
2 |
3 |
4 |
|
|
1 |
3 |
1 |
23 |
24 |
4 |
2 |
2 |
10 |
|
|
|
|
|
|
Phe |
2 |
2 |
1 |
17 |
9 |
2 |
|
4 |
1 |
1 |
|
|
|
|
1 |
2 |
|
|
|
|
|
Tyr |
|
2 |
2 |
2 |
1 |
|
3 |
2 |
2 |
4 |
|
|
1 |
1 |
|
4 |
26 |
|
|
|
|
Trp |
|
|
|
1 |
|
|
|
2 |
|
|
|
|
|
|
3 |
|
1 |
1 |
|
|
|
Pro |
5 |
35 |
5 |
4 |
1 |
|
1 |
27 |
7 |
3 |
9 |
1 |
4 |
4 |
7 |
5 |
1 |
|
|
|
This is a table of observed frequencies of amino acid replacements between closely related proteins which was indicated by Dr. Dayhoff. This table is compiled from 1572 substitutions between closely related proteins. These replacements are caused by mutations in the gene sequence. Due to the nature of the genetic code for amino acids, an amino acid residue affected by such a single genetic mutation is more likely to be replaced by some amino acids than others. The most significant deviations from the frequencies expected from random genetic mutations are shown in bold above. What is the relationship between these frequencies, and the chemical nature of the amino acid residues involved?
A:
If an amino acid mutates to another amino acids with similiar property, the three dimentional structure of this protein may not change dramatically. Absolutely, we can observe from the table that residues will mutate to that with similiar characters more frequently.Click here for more imformation
2. Why is it important to establish protein sequence homologies?
A:
If two primary sequences are more than approximately 20% identical, then they are assumed to be homologous. This homology indicates that these two sequences evolve from the same ancestor. The homology can help us to predict structure, function ,etc.
3.
(a) Define the term "chiral centre" as applied to amino acids.
A:
If Ca of an amino acid is binding to four different groups, the Ca is called a chiral centre(b) Which amino acids have chiral centres that are not alpha-carbon atoms?
A:
Ile, Thr(c) Which amino acid is this one ? Is it a D-amino acid ?
A:
Asp, L-form
4.
(a) Why the gauche(+) is the most abundant conformation of chi1?
A:
It will result to the least torsion force.(b) What is the gauche(+) conformation ?
A:
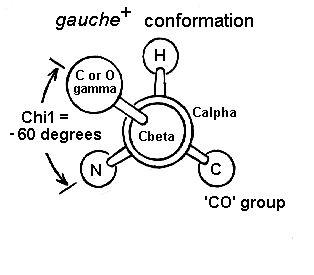
(c) Why aliphatic amino acids which are bifurcated at Cb, ie valine and isoleucine, do not adopt the trans conformation very often ?
A:
The other Cr will be in the gauche(-) position.
5.
(a ) Explain the meaning of each colored regions?
A:
White areas corresponds to comformation where atoms in the polypeptide come closer than the sum of their van der Waal radi.Red areas correspond to comformation where there are no steric clashes.
Yellow areas show the allowed regions that shorter van der Waal radi was used in caculation.
(b) Which two groups or atoms are involved for the steric hindrance in the disallowed regions of this plot?
A:
The Cb methylene group and main chain atoms are involved for the steric hindrance in the diiallowed regions.(c) Which amino acid can adopt phi and psi angles in all regions? Why?
A:
Gly,which has no side chain.
6.
(a) Why is the C-N bond length of the peptide 10% shorter than that found in usual C-N amine bonds ?
A:
The C-N bonds of a polypeptide are partial double bonds.
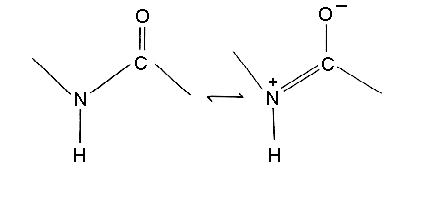
(b) Why the peptide bond nearly always has the trans configuration ? Which amino acid is found in the cis configuration more frequently than other amino acids ? Why?
A:
tras-form makes two Ca farer than that of cis-form.Pro is found in the cis-form more frequently than the other residues.
Click here for more information
7. What factors cause a-helices in globular proteins to distort ?
A:
- packing with other secondary structure
- Proline effect
- interaction with solvent